HiPoAF - Unleashing the hidden potential of anaerobic fungi (Neocallimastigomycota)

Anaerobic fungi exhibit a broad biotechnological potential. Nevertheless, there are significant knowledge gaps and many fundamental scientific questions on them, impeding methodological progress for the detection and especially maintenance of long-term cultures. We focus on basic issues in anaerobic fungi research in order to better understand their diversity, metabolic requirements and syntrophic microbial interactions. Thus, four distinct work packages were designed (https://www.hipoaf.com/).
1. Standardization (University of Innsbruck)
This work package includes a centralized media preparation and culture collection maintenance by the University of Innsbruck and their distribution among the partners.
2. Diversity and Cultivation (University of Innsbruck, Technical University of Munich)
To expand the knowledge on anaerobic fungal diversity, a database on their distribution shall be created and potential novel ecological niches for strain isolation shall be identified. Novel and known habitats shall then be screened for different representatives of Neocallimastigomycota as well as for their resting stages and potential inducers thereof. A special focus shall be laid on the identification and investigation of syntrophic interactions between anaerobic fungi and closely connected bacteria or archaea.
3. Detection and Classification (Bavarian State Research Center)
Established (qPCR, visualization) and potentially novel (FISH, live/dead staining, PCR-based techniques, metagenomics/- transcriptomics and direct RNAseq ) detection and classification techniques for anaerobic fungi in environmental as well as experimental samples shall be validated and improved. Alongside, an optimized bioinformatics workflow for taxonomic and functional analyses shall be established, that can then be used to describe and phylogenetically assign novel strains.
4. Semi-continuous Cultivation (Zurich University of Applied Sciences)
Biochemical growth requirements and cultivation parameters shall be defined with the aim to improve flow-through conditions. Therefore, suitable minimal as well as complex media for different habitats shall be defined and metabolites, growth inhibitors and promoters shall be investigated. Finally, a semi-continuous fermenter digesting lignocellulosic biomass by anaerobic fungi shall be constructed and operated.
Fungi are known to be dominant in terrestrial systems performing biological breakdown of organic carbon which is crucial for the carbon cycle. However, their role in the aquatic environment is largely uninvestigated. Different environmental and diversity studies show their presence in a brought spectrum of aquatic habitats and highlight the lack of knowledge of this kingdom. In the last years even a whole new phylum, the Cryptomycota, was discovered and proven to be present in almost every water sample taken.
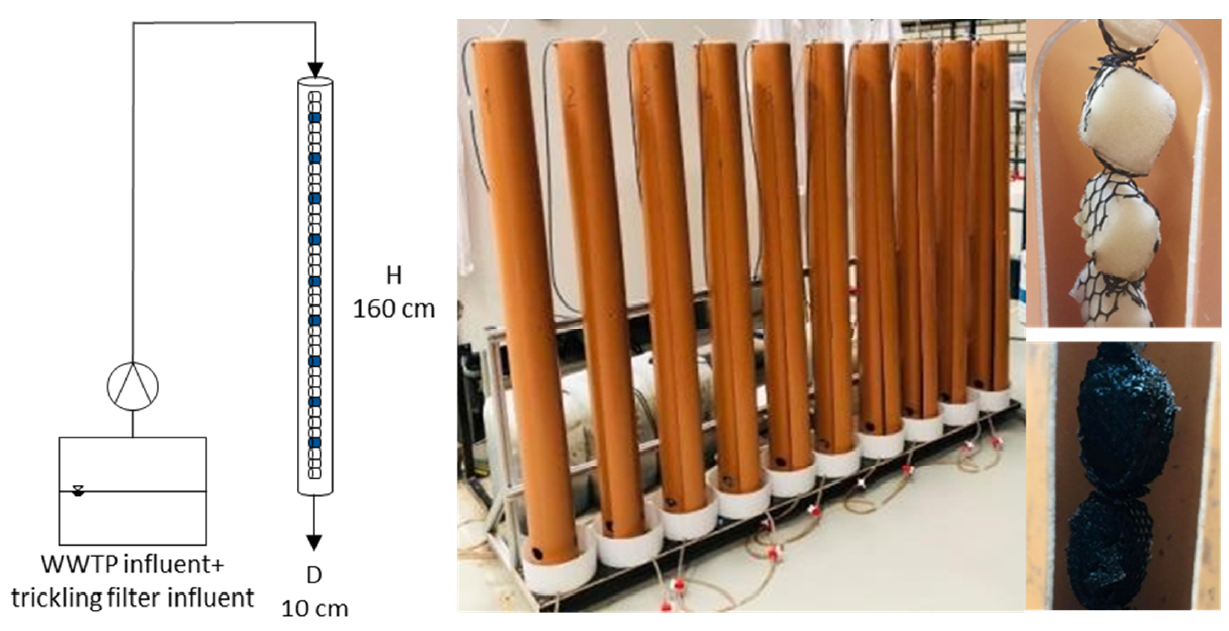
Especially in engineered biological systems, it is crucial to consider the fungal kingdom during investigations to understand and optimize the work with the whole microbial community. The superordinate objective of this research is to gain insight into and a better understanding of the fungal community in WWTPs in general with a focus on Cryptomycota. To enable this, existing molecular biological methods need to be adjusted and optimized for the use of fungi. Tackling this task this project aims to provide feasible primers for qPCR and a reliable and specific protocol. After achieving those the microbial community in down-flow hanging sponge (DHS) reactors is used as a model community to get insight into the interaction network between fungi and other microorganisms that treat wastewater. Following the influence on carbon degradation by comparing metatranscriptomic analysis of differently treated DHS reactors is studied.
A suitable qPCR system to quantify Cryptomycota was successfully implemented as well as an overview of the microbial community inside of the reactor systems was attained.








|
| |
| Project leader | Dr. Michael Lebuhn |
| Co-lead | Dr. Christian Wurzbacher |
| Responsible for the project | M.Sc. Katrin Stüer-Patowsky |
| Cooperation partner | University of Innsbruck (UIBK) Bavarian state research Center for Agriculture (LfL) Zurich University of Applied Sciences (ZHAW) Center of Biotechnology (CeBiTec) Institute of Biological, Environmental and Rural Sciences (IBERS) Institute of Animal Physiology and Genetics (IAPG) |
| Funding | Deutsche Forschungsgemeischaft (DFG) |