DMF - Exploring and identifying aquatic fungi using a combination of microscopy and next-generation molecular tools
Fungi are one important group of microorganisms that are an integral part of both terrestrial and aquatic ecosystems and play vital roles in organic matter degradation and nutrient cycling. Their diversity is estimated at 2.2 to 3.8 million species. However, many of which are still unknown so far, with only ~149 000 formally described fungal species. This disparity between known and unknown fungi is even more pronounced in aquatic habitats as only a handful of aquatic fungi are either isolated or characterized. This gets even more complicated when it comes to early diverging fungi, i.e., Microsporidia, Cryptomycota, Chytridiomycota, as there are almost no representatives in reference culture or sequence databases, and most of the described species are based on drawings. To bridge this gap between known and unknown fungi, aquatic mycologists have applied culture-independent next-generation sequencing technologies. Nevertheless, these efforts turned out to be not enough to populate the Fungal Tree of Life (FToL) with newly described fungal species.
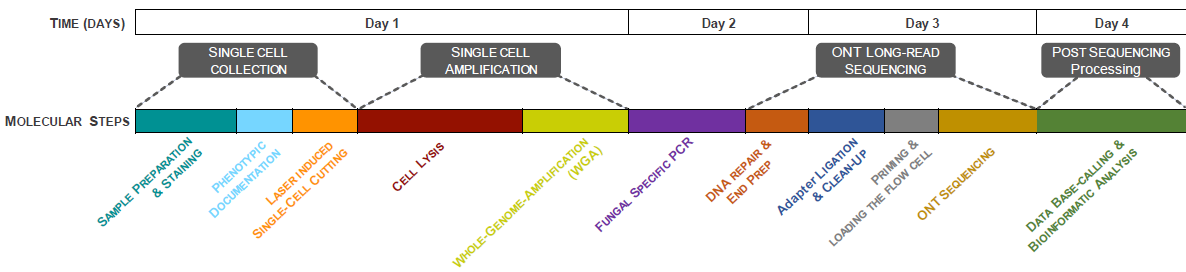
Therefore, we at the Microbial System Research Group aim to develop a toolbox to handle this key issue of Dark Matter Fungi (DMF) in aquatic habitats. For this, we are working with fungal single cells using Laser Dissection Microscope (LMD7) in combination with a third-generation long-read sequencer (Oxford Nanopore) (Figure 1). By combining laser dissection of fungal single cells with complete fungal ribosomal operon amplification, we are establishing a methodological platform (Eco-ACDCS: Ecological Annotated Cell Dissection for Cultivation and nucleic acid Sequencing) which will be the key to resolve the fungal dark matter in aquatic habitats.
With the realization of the Eco-ACDCS platform, we will systematically screen and catalog the unknown fungal lineages in our environment with the aim to:
· grasp the identity of aquatic fungi, in particular, when working with DNA sequence data
· resolve the phylogeny of novel fungal lineages by using multiple markers or by moving to single cells
· perform high-throughput analysis of freshwater fungi from varying environmental samples
· bridge between the past and the future in providing references for aquatic fungi
· provide a referenced DNA bank for single cells and subsequent genome analysis
With the Eco-ACDCS platform, we will play a significant role in driving the aquatic mycology from unknowns to knowns, especially for early diverging fungal lineages. Our reference sequences and the DNA bank of fungal single cells will further facilitate future research.

Therefore, being part of the Microbial System Research Group, we have successfully established a workflow that combines laser microdissection of fungal single cells with whole genome amplification and long-read sequencing. We have tested different aquatic habitats for hunting of specific fungal groups with success, e.g., aquatic hyphomycetes (known for leaf litter degradation) and chytrids (parasitic fungi). We have achieved a success rate of 59% for fugal short ITS region amplification and 24% for longer ribosomal region (5kb) of fungal rDNA. This workflow has enabled us to identify the taxonomic and phylogenic placement of an unknown fungal species within a timeframe of 1 week (Figure 2).
Moreover, we have expanded the application of our workflow to the aquatic habitats of the Arctic regions of the Northern Canada and the extreme water bodies of Iceland. During summer 2022, we have carried out sampling visits to the Arctic field sites of Northern Canada to sample permafrost thaw ponds of different ages (Figure 41a) and the glacial lakes and thermal hot springs of Iceland (Figure 41b). The samples will be processed and will help to tap the unknown fungal lineages from such environments which are critical to study from the perspective of global warming and extremophilic mycology.

|
| |
| Project leader | Dr. Christian Wurzbacher |
| Responsible for the project | |
| Funding | Deutsche Forschungsgemeinschaft - DFG |


